=================================================================================
Using Variance Inflation Factors (VIFs) is an effective way to investigate multicollinearity in a multiple regression model. VIFs measure how much the variance of the estimated regression coefficients is increased due to multicollinearity. Higher VIF values suggest stronger multicollinearity. The VIF for the j-th variable is calculated as:
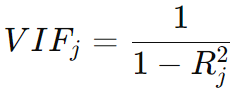 -------------------------------------------- [4005a] -------------------------------------------- [4005a]
where Rj2 is the coefficient of determination of a regression of Xj on all the other predictors. A VIF value greater than 10 is often taken as an indication that multicollinearity may be unduly influencing the model..
Here's how you can investigate multicollinearity and assess the impact of removing BMI from the model in JMP:
-
Calculate VIFs for the Full Model:
- Fit the full regression model with "%Fat" as the response and all predictors, including BMI.
- Right-click on the "Parameter Estimates" table.
- Select "Columns" and then choose "VIF" to add VIF values to the table.
- Assess VIF Values:
- Examine the VIF values for each predictor variable in the full model.
- Typically, VIF values above 10 or 5 are considered indicative of significant multicollinearity. Lower values are better.
- Remove BMI from the Model:
- Remove the BMI variable from the predictor list and refit the regression model.
- Calculate VIFs for the Reduced Model:
- Right-click on the "Parameter Estimates" table for the reduced model.
- Again, select "Columns" and then choose "VIF" to add VIF values to the table for the reduced model.
- Compare VIFs:
- Compare the VIF values for each predictor variable in the full model (with BMI) and the reduced model (without BMI).
- Assess whether removing BMI has a notable impact on the VIF values for the other predictors.
In general, if the VIF values for all predictors in the reduced model decrease significantly after removing BMI, it suggests that BMI was contributing to multicollinearity in the model. Lower VIF values in the reduced model indicate reduced multicollinearity.
However, it's important to keep in mind that multicollinearity is a relative concept, and the specific threshold for what is considered problematic multicollinearity may vary depending on the context and goals of your analysis. Therefore, you should consider both the magnitude of the VIF values and the practical implications of multicollinearity in your interpretation of the results.
Variance Inflation Factors (VIFs) are useful for detecting multicollinearity in yield analysis within the semiconductor industry. Here are some examples of how VIFs can be applied:
-
Process Parameters: In semiconductor manufacturing, various process parameters, such as temperature, pressure, and gas flow rates, can affect yield. You can use VIFs to assess whether these process parameters are highly correlated. For example, if VIF values for temperature and pressure are both high, it indicates multicollinearity, suggesting that changes in one parameter may be associated with changes in the other.
-
Equipment Settings: Semiconductor production involves different equipment settings, like deposition rates, tool power levels, and etch process parameters. VIFs can help you identify whether these equipment settings are highly correlated. If several equipment settings have high VIFs, it suggests that they may be interrelated and could lead to multicollinearity in the yield analysis.
-
Material Properties: The semiconductor industry relies on various materials with different properties. You can use VIFs to investigate whether material properties like resistivity, dielectric constant, and thermal conductivity are correlated. High VIFs for material properties indicate multicollinearity, which can affect yield predictions.
-
Production Steps: Semiconductor manufacturing involves a sequence of production steps, such as lithography, etching, and chemical vapor deposition. VIFs can help determine if these production steps are highly correlated. For example, if VIFs for two specific production steps are high, it suggests that they may influence each other, leading to multicollinearity.
-
Environmental Factors: Factors like humidity and air quality can impact semiconductor manufacturing processes and yield. VIFs can be used to investigate whether these environmental factors are highly correlated. If VIFs for environmental factors are elevated, it implies that they may exhibit multicollinearity in yield analysis.
-
Defect Characteristics: In yield analysis, you might consider various defect characteristics, such as defect size, defect density, and defect type. VIFs can help identify whether these defect characteristics are highly correlated. High VIFs in this context may indicate that certain defect characteristics tend to co-occur, potentially affecting yield.
By calculating VIFs for these variables and assessing their magnitudes, semiconductor engineers and analysts can gain insights into the presence and severity of multicollinearity in yield analysis. Addressing multicollinearity through variable selection, transformation, or other techniques can help improve the accuracy and interpretability of yield models, leading to better decision-making in semiconductor manufacturing.
============================================
|
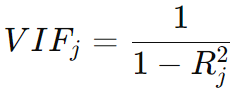 -------------------------------------------- [4005a]
-------------------------------------------- [4005a]